Data-Driven Discovery
Genevera’s group develops new statistical machine learning tools to help people make discoveries from large and complex data sets. View Full List of Publications on Google ScholarSTATISTICAL MACHINE LEARNING

Graphical Models
We develop new types of probabilistic graphical models and graph learning strategies for representing, discovering, and visualizing relationships in large data sets. Our work includes developing new classes of graphical models for diverse data types as well as mixed or multi-modal data. A recent focus of our work is on developing graph learning strategies for large-scale neuroscience data.
Key Publications:
- E. Yang, P. Ravikumar, G. I. Allen, and Z. Liu, “Graphical Models via Univariate Exponential Family Distributions”, Journal of Machine Learning Research, 16, 3813-3847, 2015.
- E. Yang, Y. Baker, P. Ravikumar, G. I. Allen, and Z. Liu, “Mixed Graphical Models via Exponential Families”, Artificial Intelligence and Statistics (AISTATS), oral presentation, 2014.
- E. Yang, P. Ravikumar, G. I. Allen, and Z. Liu, “On Poisson Graphical Models”, In Advances in Neural Information Processing Systems (NeurIPS), 2013.
- G. Vinci, G. Dasarathy, and G. I. Allen, “Graph Quilting: Graphical Model Selection from Partially Observed Covariances”, (Submitted), arXiv:1912.05573, 2022.
- M. Wang and G. I. Allen, “Thresholded graphical lasso adjusts for latent variables“, (Submitted), arXiv:2104.06389, 2022.
Dimension Reduction
Dimension reduction techniques are used for visualizing, exploring, and discovering patterns in large data sets. We have developed many dimension reduction techniques for complex and structured data; these include sparse tensor decompositions and generalizations of PCA for structured or multi-modal data.
Key Publications:
- G. I. Allen, L. Grosenick and J. Taylor, “A Generalized Least Squares Matrix Decomposition”, Journal of the American Statistical Association: Theory and Methods, 109:505, 145-159, 2014.
- G. I. Allen, “Sparse Higher-Order Principal Components Analysis”, Artificial Intelligence and Statistics (AISTATS), 2012.
- T. M. Tang and G. I. Allen, “Integrated Principal Components Analysis”, Journal of Machine Learning Research, 22:84, 1-71, 2021.
- G. I. Allen, and M. Weylandt, “Sparse and Functional Principal Components Analysis”, In Proceedings of the IEEE Data Science Workshop, 2019.
Clustering
Clustering seeks to find groups in large data sets. We have developed several convex clustering approaches that offer accurate, principled, and flexible strategies along with built-in visualizations for clustering.
Key Publications:
- M. Weylandt, J. Nagorski, and G. I. Allen, “Dynamic Visualization and Fast Computation for Convex Clustering via Algorithmic Regularization”, Journal of Computational and Graphical Statistics, 29:1, 87-96, 2020.
- E. C. Chi, G. I. Allen, and R. Baraniuk, “Convex Biclustering”, Biometrics, 73:1, 10-19, 2017.
- M. Wang and G. I. Allen, “Integrative Generalized Convex Clustering Optimization and Feature Selection for Mixed Multi-View Data”, Journal of Machine Learning Research, 22:1:73, 2021.
- M. Weylandt, TM Roddenberry, G. I. Allen, “Simultaneous Grouping and Denoising via Sparse Convex Wavelet Clustering”, In IEEE Data Science & Learning Workshop (DSLW), arXiv:2012.04762, 2021.
Feature Selection
We develop a variety of feature selection techniques to improve the interpretability of machine learning methods. Our specific goal is to select relevant features from highly correlated and high-dimensional data sets.
Key Publications:
- F. Campbell and G. I. Allen, “Within Group Variable Selection through the Exclusive Lasso”, Electronic Journal of Statistics, 11:2, 4220-4257, 2017.
- G. I. Allen, “Automatic Feature Extraction via Weighted Kernels and Regularization”, Journal of Computational and Graphical Statistics, 22:2, 284-299, 2013.
- Y. Baker, T. M. Tang, and G. I. Allen, “Feature Selection for Data Integration with Mixed Multiview Data”, Annals of Applied Statistics, 14:4, 1676-1698, 2020.
- T. Yao and G. I. Allen, “Feature Selection for Huge Data via Minipatch Learning”, (Submitted), arXiv:2010.08529, 2022.
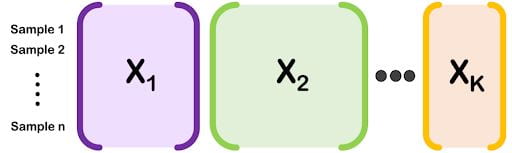
Data Integration
Large data sets are often diverse, with multiple types of features measured on the same set of subjects or observations. We have developed a variety of interpretable machine learning techniques for discovering joint patterns in this so-called mixed multi-modal data.
Key Publications:
- E. Yang, Y. Baker, P. Ravikumar, G. I. Allen, and Z. Liu, “Mixed Graphical Models via Exponential Families”, Artificial Intelligence and Statistics (AISTATS), oral presentation, 2014.
- Y. Baker, T. M. Tang, and G. I. Allen, “Feature Selection for Data Integration with Mixed Multiview Data”, Annals of Applied Statistics, 14:4, 1676-1698, 2020.
- M. Wang and G. I. Allen, “Integrative Generalized Convex Clustering Optimization and Feature Selection for Mixed Multi-View Data”, Journal of Machine Learning Research, 22:1:73, 2021.
- T. M. Tang and G. I. Allen, “Integrated Principal Components Analysis”, Journal of Machine Learning Research, 22:84, 1-71, 2021.
Ensemble Learning
Recently, we have begun developing new computationally efficient ensemble learning strategies that also lead to improved accuracy and interpretability.
Key Publications:
- Y. Hu and G. I. Allen, “Local-Aggregate Modeling for Big-Data via Distributed Optimization: Applications to Neuroimaging”, Biometrics, 71:4, 905-917, 2015.
- T. Yao and G. I. Allen, “Feature Selection for Huge Data via Minipatch Learning”, (Submitted), arXiv:2010.08529, 2022.
- M. T. Toghani and G. I. Allen, “MP-Boost: Minipatch Boosting via Adaptive Feature and Observation Sampling”, In IEEE International Conference on Big Data and Smart Computing (BigComp), 2021.
Interpretability & Fairness
We recently have begun work on machine learning ethics related to interpretability and algorithmic fairness. Our goal is to develop principled approaches for reliably interpreting or unraveling black-box machine learning approaches as well as mitigating biases in machine learning predictions.
Key Publications:
- L. Gan*, L. Zheng* and G. I. Allen, “Fast, Model-Agnostic Confidence Intervals for Feature Importance“, (Submitted) arXiv:2206.02088, 2022. (*Denotes equal contribution.)
- C. O. Little*, M. Weylandt and G. I. Allen, “To the Fairness Frontier and Beyond: Identifying, Quantifying, and Optimizing the Fairness-Accuracy Pareto Frontier“, (Submitted) arXiv:2206.00074, 2022 (*Denotes equal contribution.)
APPLICATIONS
Neuroscience
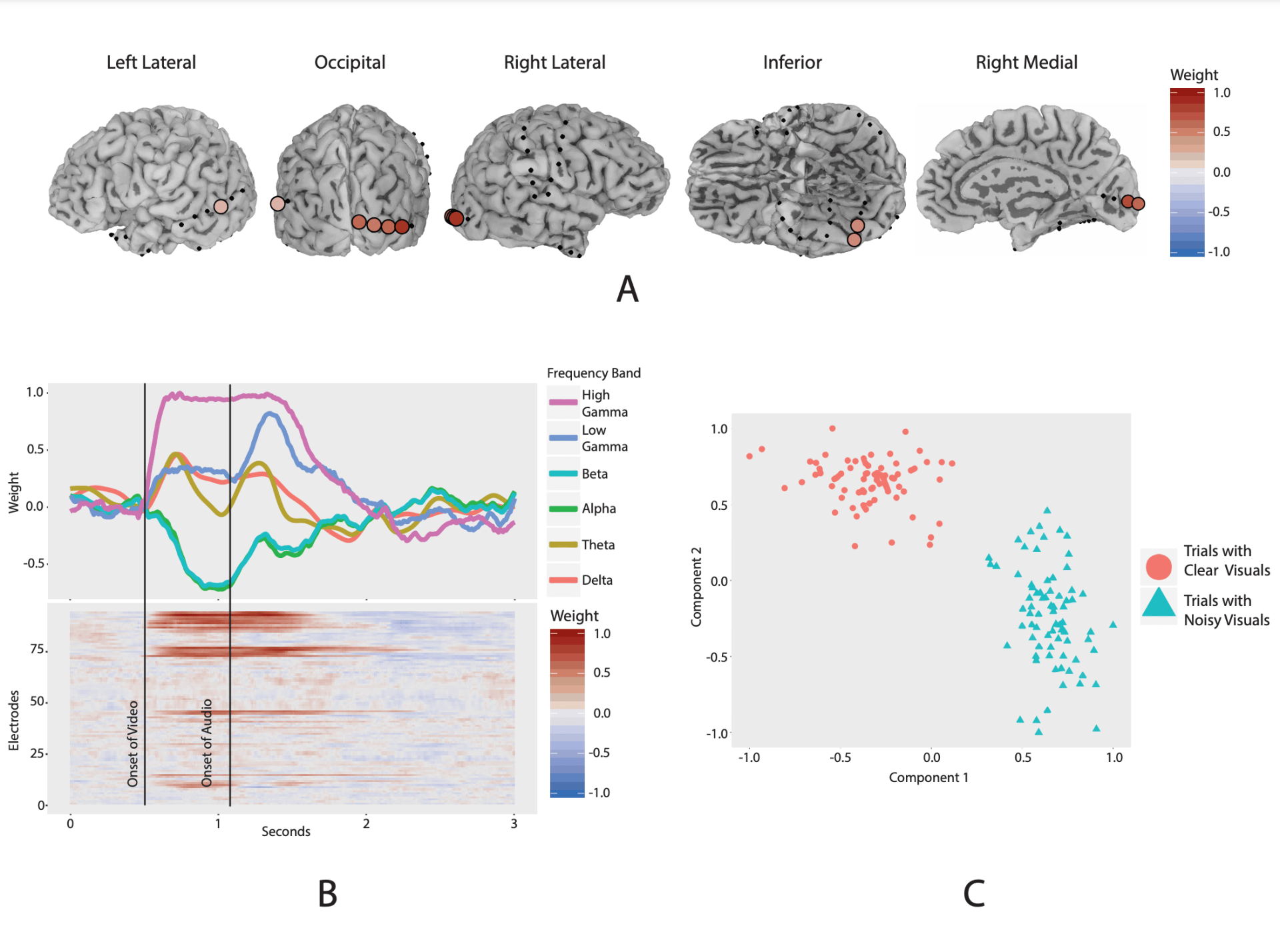
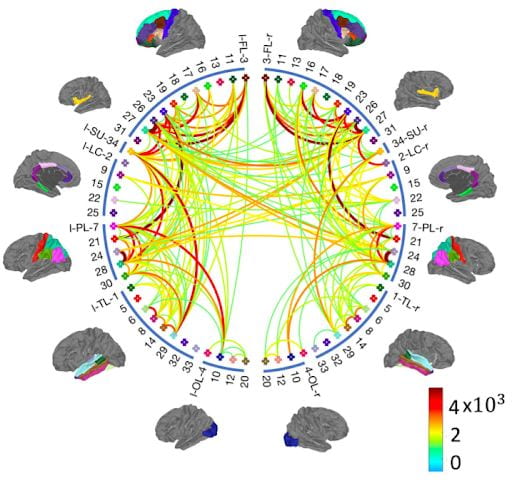
We develop new statistical machine learning approaches to analyze huge data sets from new technologies for neuroimaging and neural recordings. A key goal of our research is to understand brain connectomics, or how the brain is functionally and structurally connected.
Key Publications:
- S. Tomson, M. Narayan, G. I. Allen, D. Eagleman, “Neural Networks of Synesthesia”, Journal of Neuroscience, 33:35, 14098-14106, 2013.
- Z. Zhang, G. I. Allen, H. Zhu, D. Dunson, “Tensor Network Factorizations: Relationships between Human Brain Structural Connectomes and Traits”, NeuroImage, 197:330- 343, 2019.
- M. Narayan and G. I. Allen, “Mixed Effects Models for Resampled Network Statistics Improves Statistical Power to Find Differences in Multi-Subject Functional Connectivity” Frontiers in Neuroscience, 10:108, 2016.
- K. Geyer, F. Campbell, A. Chang, J. Magnotti, M. Beauchamp, G. I. Allen, “Interpretable Visualization and Higher-Order Dimension Reduction for ECoG Data”, In Proceedings of the International Workshop on Big Data Reduction held with the 2020 IEEE International Conference on Big Data, 2020.
- A. Chang, T. Yao, and G. I. Allen, “Graphical Models and Dynamic Factor Models for Modeling Functional Brain Connectivity”, In Proceedings of the IEEE Data Science Workshop, 2019.
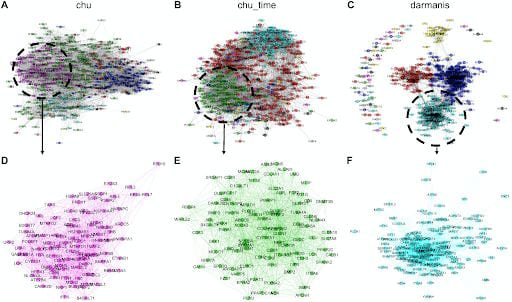
Bioinformatics
New biomedical technologies have led to an enormous proliferation of “omics” data measuring DNA, RNA, proteins, metabolites and more. We develop new statistical machine learning techniques to make discoveries from high-dimensional omics data as well as data integration techniques for analyzing multi-omics data.
Key Publications:
- Y. W. Wan, G. I. Allen, and Z. Liu, “TCGA2STAT: Simple TCGA Data Access for Integrated Statistical Analysis in R”, Bioinformatics, 32:6, 952-954, 2016.
- G. I. Allen and Z. Liu, “A Local Poisson Graphical Model for Inferring Networks from Next Generation Sequencing Data”, IEEE Transactions on NanoBioscience, 12:3, 1-10, 2013.
- G. I. Allen and M. Maletic-Savatic, “Sparse Non-negative Generalized PCA with Applications to Metabolomics”, Bioinformatics, 27:21, 3029-3035, 2011.
- L. Gan, G. Vinci, and G. I. Allen, “Correlation Imputation in Single-Cell TNA-Seq using Auxiliary Information and Ensemble Learning”, In Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, 1-6, 2020.
We gratefully acknowledge support from:

